COMPETITIVE EXAM MCQs SERIES of LIFE SCIENCES for CSIR-UGC NET/JRF, SLET, GATE, and other entrance tests – MOLECULES AND THEIR INTERACTION RELEVANT TO BIOLOGY – Protein and DNA Stability Factors.
Syllabus Outline
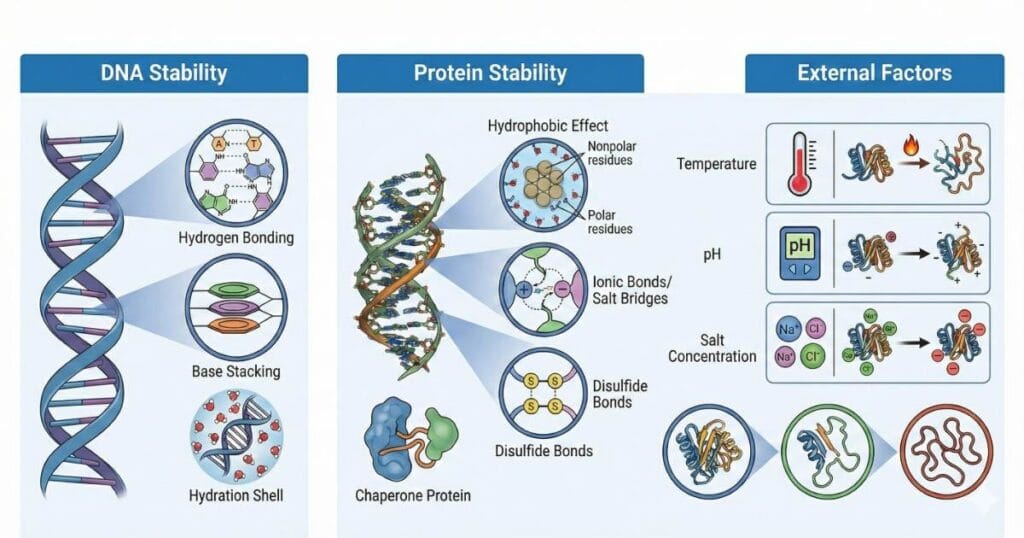
- Factors affecting protein stability – pH, temperature, solvents
- Denaturation and renaturation of proteins
- Protein folding landscape and chaperonins
- DNA melting temperature (Tm) and GC content
- Nucleic acid denaturation and hybridisation
- Role of salt, pH, and organic solvents in stability
- Stabilising interactions – hydrogen bonds, disulfide bonds
- Mutations affecting stability and function
This quiz contains concept-based, most frequently asked 25 MCQs of “MOLECULES AND THEIR INTERACTION RELEVANT TO BIOLOGY – Protein and DNA Stability Factors”. Each question has a single correct/most appropriate answer.
*****
1. In a protein engineering study, a Glycine residue in a solvent-exposed loop is mutated to Alanine. This mutation increases the protein’s Tm. What is the thermodynamic basis for this stabilisation?
A) New hydrogen bonds formed by the Alanine methyl group.
B) Reduction in the conformational entropy of the unfolded state
C) Increase in the enthalpy of the folded state due to better packing.
D) Removal of a destabilising electrostatic clash.
2. Cold denaturation of proteins occurs because:
A) The entropy of the polypeptide chain becomes zero at low temperatures.
B) Water crystallises in the hydrophobic core, disrupting van der Waals interactions.
C) The hydrophobic effect weakens at low temperature, and non-polar groups become hydrated.
D) Hydrogen bonds become weaker as the temperature decreases.
3. The large positive heat capacity change observed upon protein unfolding is linearly correlated with:
A) The disruption of hydrogen bonds in the alpha-helices.
B) The increase in vibrational entropy of the backbone.
C) The number of disulfide bonds broken.
D) The change in Solvent Accessible Surface Area of non-polar residues.
4. Reducing the disulfide bonds of a protein destabilises the native state primarily by:
A) Decreasing the enthalpy of the native state.
B) Increasing the conformational entropy of the unfolded state.
C) Reducing the heat capacity change of unfolding.
D) Increasing the solubility of the unfolded state.
5. The “Iterative Annealing” mechanism proposes that GroEL facilitates folding by:
A) Actively bending the polypeptide into the native shape using hydrophobic interactions.
B) Unfolding kinetically trapped misfolded intermediates
C) Providing a template sequence for the protein to copy.
D) Increasing the local concentration of ATP near the substrate.
6. How does “Molecular Crowding” in the cellular environment primarily affect protein folding compared to dilute in vitro conditions?
A) It stabilises the unfolded state by providing more excluded volume.
B) It increases the association constant for intermolecular aggregation
C) It reduces the viscosity of the solvent, accelerating folding rates.
D) It reduces the concentration of water, which eliminates hydrophobic interaction.
7. Increasing the concentration of NaCl from 10 mM to 100 mM raises the melting temperature (Tm) of DNA. This is best explained by:
A) Increased hydrophobic bonding between bases.
B) Transition from B-DNA to A-DNA due to low water concentration.
C) Formation of sodium bridges between base pairs.
D) Reduce repulsion by shielding phosphates on DNA.
8. Which interaction makes the largest enthalpic contribution to the stability of the DNA double helix?
A) Hydrogen bonding between base pairs.
B) Ionic interactions with solvent.
C) Phosphate-sugar backbone rigidity.
D) Base stacking interactions.
9. The “Hyperchromic Effect” observed during DNA denaturation is caused by:
A) The ionisation of bases at high temperature.
B) The loss of base stacking interactions
C) The hydrolysis of the phosphodiester backbone.
D) Water molecules are released from the major groove.
10. Upon unfolding of a protein, the fluorescence emission maximum of a buried Tryptophan residue shifts from 330 nm to 355 nm. This “Red Shift” occurs because:
A) The residue moves into an aqueous environment.
B) The residue forms an excimer with a tyrosine.
C) The quantum yield of the tryptophan increases in water.
D) The residue moves into a more hydrophobic environment.
11. A protein state that retains native-like secondary structure but lacks fixed tertiary packing and binds hydrophobic dyes is called:
A) The Amyloid state.
B) The Molten Globule state.
C) The Transition state.
D) The Native state.
12. Assertion (A): The refolding kinetics of Proline-containing proteins often exhibit a very slow phase.
Reason (R): The peptide bond preceding Proline can exist in a cis configuration, and cis-trans isomerisation is intrinsically slow.
A) Both A and R are correct, and R is the correct explanation of A.
B) Both A and R are correct, but R is not the explanation of A.
C) A is correct, but R is incorrect.
D) A is incorrect, but R is correct.
13. Arrange the following nucleic acid duplexes in decreasing order of stability in a high ionic strength buffer:
I – DNA : DNA
II – RNA : RNA
III – RNA : DNA
A) I > II > III
B) II > III > I
C) III > I > II
D) II > I > III
14. Why is RNA susceptible to hydrolysis in alkaline conditions while DNA is stable?
A) Uracil is more acidic than Thymine.
B) The 2′-OH group of ribose acts as a nucleophile, attacking the phosphodiester bond.
C) DNA forms a double helix, which protects the backbone, while RNA is single-stranded.
D) RNA contains weaker glycosidic bonds.
15. A DNA sample has an A260/A280 ratio of 1.2. This indicates:
A) The sample is pure DNA.
B) The sample is contaminated with protein.
C) The sample is contaminated with RNA.
D) The sample contains phenol.
16. Trifluoroethanol is often used to stabilise alpha-helices in peptides. Its mechanism of action involves:
A) Acting as a strong hydrogen bond donor to the peptide backbone.
B) Strengthening intramolecular H-bonds
C) Binding to the N-terminus of the helix.
D) Increasing the viscosity of the solvent.
17. When calculating the Tm of a short oligonucleotide probe, the “Nearest Neighbour” method is more accurate than the simple Wallace rule because:
A) It accounts for the length of the primer.
B) It considers the sequence-dependent stacking energy.
C) It considers the salt concentration to calculate Tm in addition to the sequence.
D) It assumes all G-C/A-T pairs have identical stability.
18. Assertion (A): Linear DNA has a higher Tm than covalently closed circular DNA of the same sequence.
Reason (R): Unwinding of the helix in linear DNA introduces positive supercoiling, which is energetically unfavourable and resists denaturation.
A) Both A and R are correct, and R is the correct explanation of A.
B) Both A and R are correct, but R is not the explanation of A.
C) A is incorrect, but R is correct.
D) Both A and R are incorrect.
19. Fluorescence Anisotropy is a technique used to measure protein binding. Upon binding to a large receptor, the anisotropy of a small fluorescent ligand:
A) Decreases because the ligand rotates faster.
B) Decreases due to quenching.
C) Remains unchanged.
D) Increases because the effective molecular weight increases
20. Which combination of forces favours the folded state of a globular protein?
I – Conformational entropy of the chain.
II – Hydrophobic effect.
III – Intramolecular Hydrogen Bonding.
IV – Van der Waals interactions in the core.
A) I, II and III
B) I, II, III and IV
C) I and IV only
D) II, III and IV
21. Assertion (A): Levinthal’s paradox states that a protein cannot fold by randomly searching all possible conformations.
Reason (R): The time required for a random search would exceed the age of the universe.
A) Both A and R are correct, and R is the correct explanation of A.
B) Both A and R are correct, but R is not the explanation of A.
C) A is correct, but R is incorrect.
D) A is incorrect, but R is correct.
22. How does Sodium Dodecyl Sulfate denature proteins for SDS-PAGE?
A) It reduces disulfide bonds.
B) It cleaves peptide bonds at tryptophan residues.
C) It unfolds the protein and imparts a uniform negative charge.
D) It cross-links Lysine residues.
23. The addition of Ethidium Bromide (EtBr) to DNA increases its melting temperature (Tm). This is because:
A) EtBr acts as a condensing agent.
B) Intercalation of EtBr stabilises the double helix.
C) EtBr forms covalent cross-links between strands.
D) EtBr neutralises the backbone charge.
24. The linking number of a closed circular DNA is defined as:
I – Linking Number = Twist + Writhe
II – Linking Number is an integer for closed circles.
III – Linking Number changes when DNA bends.
IV – Linking Number changes by breaking the backbone.
Which statements are correct?
A) I and II only
B) I, II and IV
C) I, III and IV
D) II and III only
25. Assertion (A): Anfinsen’s dogma applies to all proteins without exception.
Reason (R): Some proteins, like prions or metamorphic proteins, can adopt multiple stable conformations from the same sequence depending on the environment or template.
A) Both A and R are correct.
B) A is correct, and R is incorrect.
C) A is incorrect, and R is correct.
D) Both A and R are incorrect.
*****
Previous: Nucleic Acids Structure and Conformation
Next: Carbohydrate and Lipid Metabolism
References
- Nelson, David L. & Cox, Michael M. (2021). Lehninger Principles of Biochemistry, W. H. Freeman, 8th Edition
- Voet, Donald, Voet, Judith G., & Pratt, Charlotte W. (2018). Voet’s Principles of Biochemistry, Wiley, 5th Edition
- Berg, Jeremy M., Tymoczko, John L., & Stryer, Lubert (2023). Biochemistry, W. H. Freeman, 10th Edition
- Palmer, Trevor & Bonner, Philip L. (2007). Enzymes: Biochemistry, Biotechnology, Clinical Chemistry, Horwood Publishing, 2nd Edition
- Upadhyay, Avinash, Upadhyay, K., & Nath, Nirmalendu (2023). Biophysical Chemistry: Principles and Techniques, Himalaya Publishing House, 4th Edition
🔗 Explore More MCQs: